The Endpoint
Chemical substances, mixtures of substances and physical agents (e.g., radiation) can induce alterations in the genome of either somatic or germinal cells. There are several mutagenic endpoints of concern; these include point mutations (i.e., submicroscopic changes in the base sequence of DNA) and structural or numerical chromosome aberrations. Structural aberrations include deficiencies, duplications, insertions, inversions, and translocations, whereas numerical aberrations are gains or losses of whole chromosomes (e.g., trisomy, monosomy) or sets of chromosomes (haploidy, polyploidy).
Certain mutagens, such as alkylating agents, can directly induce alterations in the DNA. Mutagenic effects may also come about through mechanisms other than chemical alterations of DNA. Among these are interference with normal DNA synthesis (as caused by some metal mutagens), interference with DNA repair, abnormal DNA methylation, abnormal nuclear division processes, or lesions in non-DNA targets. Evidence that an agent induces heritable mutations in human beings could be derived from epidemiologic data indicating a strong association between chemical exposure and heritable effects. It is difficult to obtain such data because any specific mutation is a rare event, and only a small fraction of the estimated thousands of human genes and conditions are currently useful as markers in estimating mutation rates.
Mutagenicity & REACH
Mutagenicity studies are required for all the tonnage bands. Unlike most other endpoints, a negative mutagenicity result in vitro can be considered sufficient evidence for non-mutagenic potential but positive results must be confirmed in vivo. At 1-10 tonnage level (Annex VII) the in vitro gene mutation study in bacteria (Ames test) is required. At Annex VIII level (10-100 ton), two additional in vitro studies are required: a cytogenicity study and a gene mutation study in mammalian cells. If there is any positive result within these in vitro tests, in vivo mutagenicity studies are required. At higher tonnage in vivo studies are needed.
The Model


This model has been implemented in the CAESAR freeware
(both on-line and stand-alone versions are available)
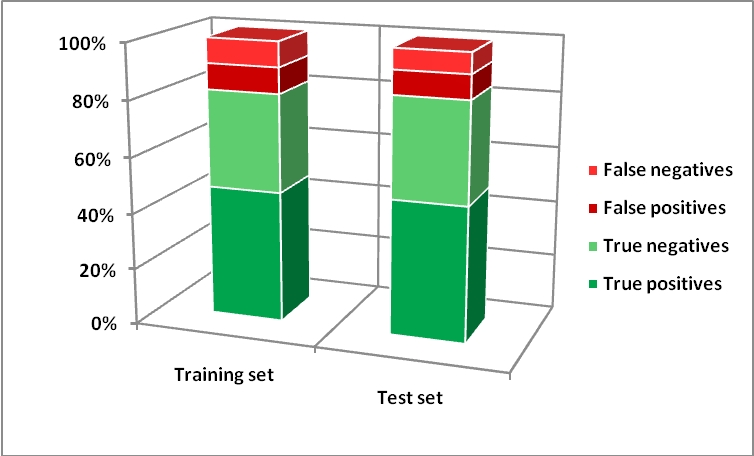
The CAESAR model for mutagenicity is based on a data set that includes 4225 compounds. For developing classification models, this data set was subdivided in two classes: 80% (3380 chemicals) used for building the model and 20% (845 chemicals) left for testing.
For regulatory purposes, an integrated model was arranged cascading two complementary techniques: a machine learning algorithm (SVM), to build an early model with the best statistical accuracy, equipped with an expert facility for FN removal based on known structural alerts, to refine its predictions.
The figure below shows results obtained with SVM:
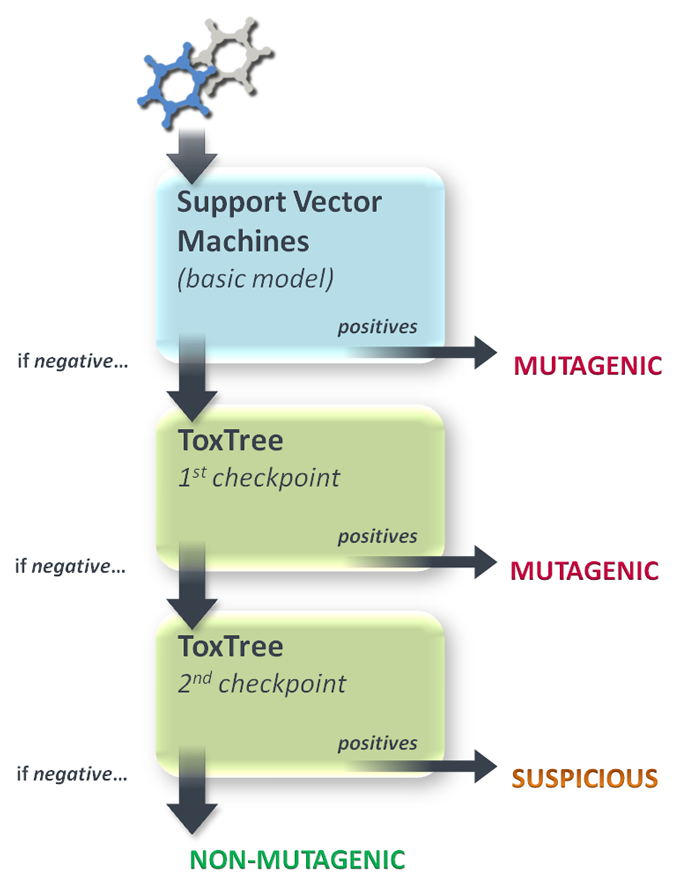
The expert filter, to be applied only on compounds presumed safe by SVM, wraps two set of SA (selected from the Benigni/Bossa rulebase) with different distinguishing features: the former (the 'sharp' one) has the aim to enhance the prediction accuracy attempting a precise identification of misclassified FN, the latter (the 'suspicious' one) goes on with the FN removal as much as this doesn't noticeably downgrade the original prediction accuracy (by generating too many FP as well). To point out this distinction, compounds picked out by the first checkpoint are classified as 'mutagenic', and those picked out by the second are classified as 'suspicious'. Unaffected ones are finally classified as 'non-mutagenic'. The figure below shows the overall scheme:
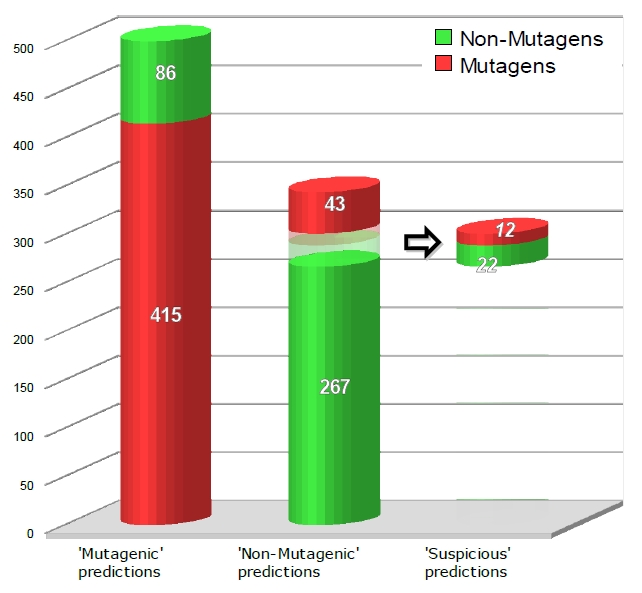
A global overlook of the performances of the combined model on the test set is illustrated in figure below, where a nice interpretation of the 'suspicious' rules set is given: it can extract from the presumed safe compounds the most suspect ones with an impressive specificity, if related to the very low number of real mutagens still present.
References
- An open source multistep model to predict mutagenicity from statistical analysis and relevant structural alerts.
Ferrari T., Gini G.
Chemistry Central Journal 2010, 4(Suppl 1):S2 (29 July 2010)